Foundational Questions for Research in the Shilatifard Lab
Foundational Questions
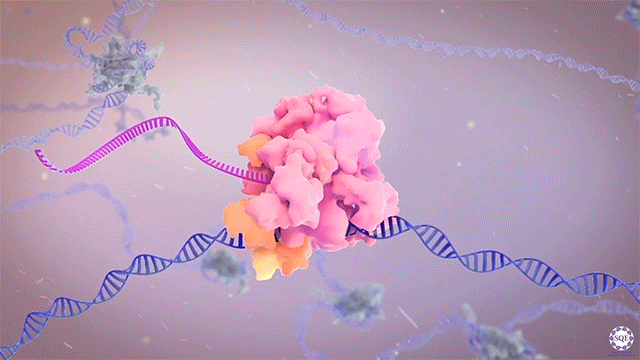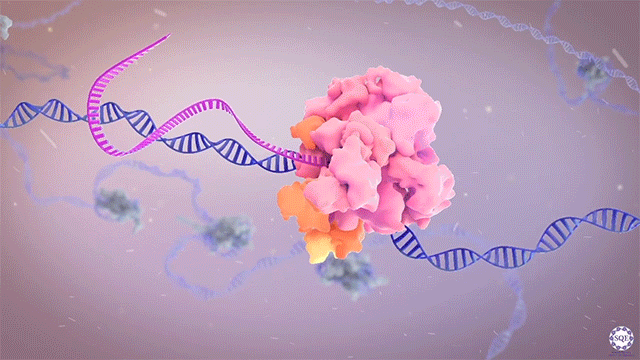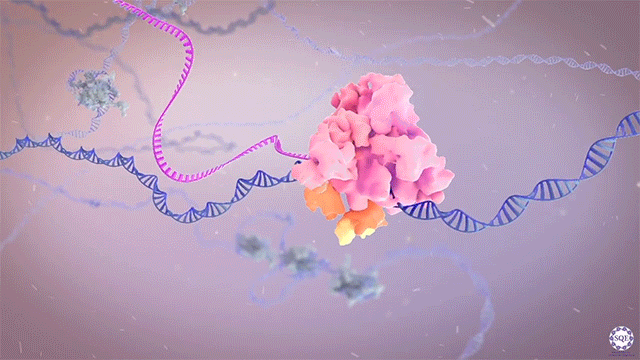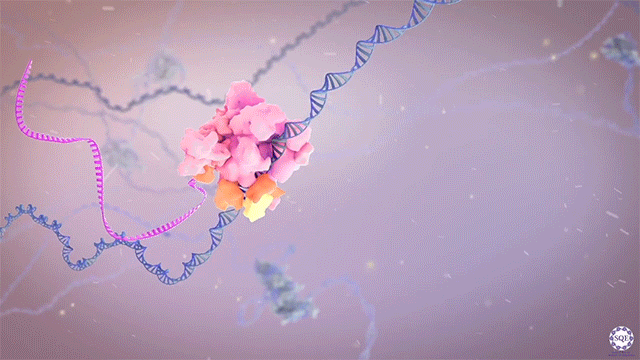
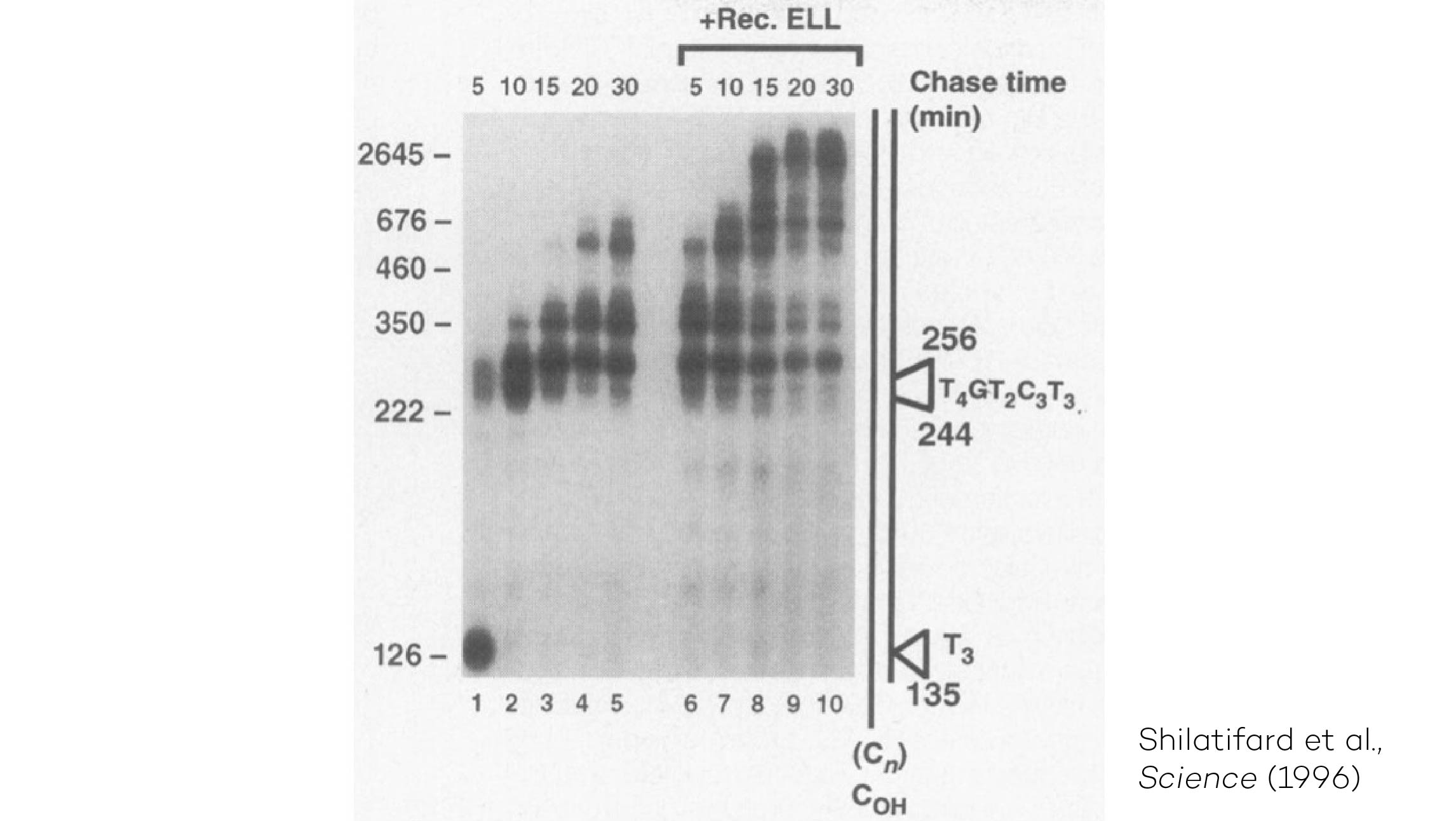
The Shilatifard laboratory’s decades-long history of contributions to transcriptional regulation, epigenetics, chromatin biology and their link to human disease originates from a single question: why do translocations of the MLL gene on chromosome 11 into so many seemingly unrelated genes on different chromosomes all result in oncogenesis of acute leukemia? We arrived at this question in 1996, when we identified the product of the human gene ELL, a known MLL translocation partner, as a transcriptional elongation factor. This finding was the first reported link between human cancer and transcriptional elongation control, and it began a lifelong scientific journey for our laboratory.
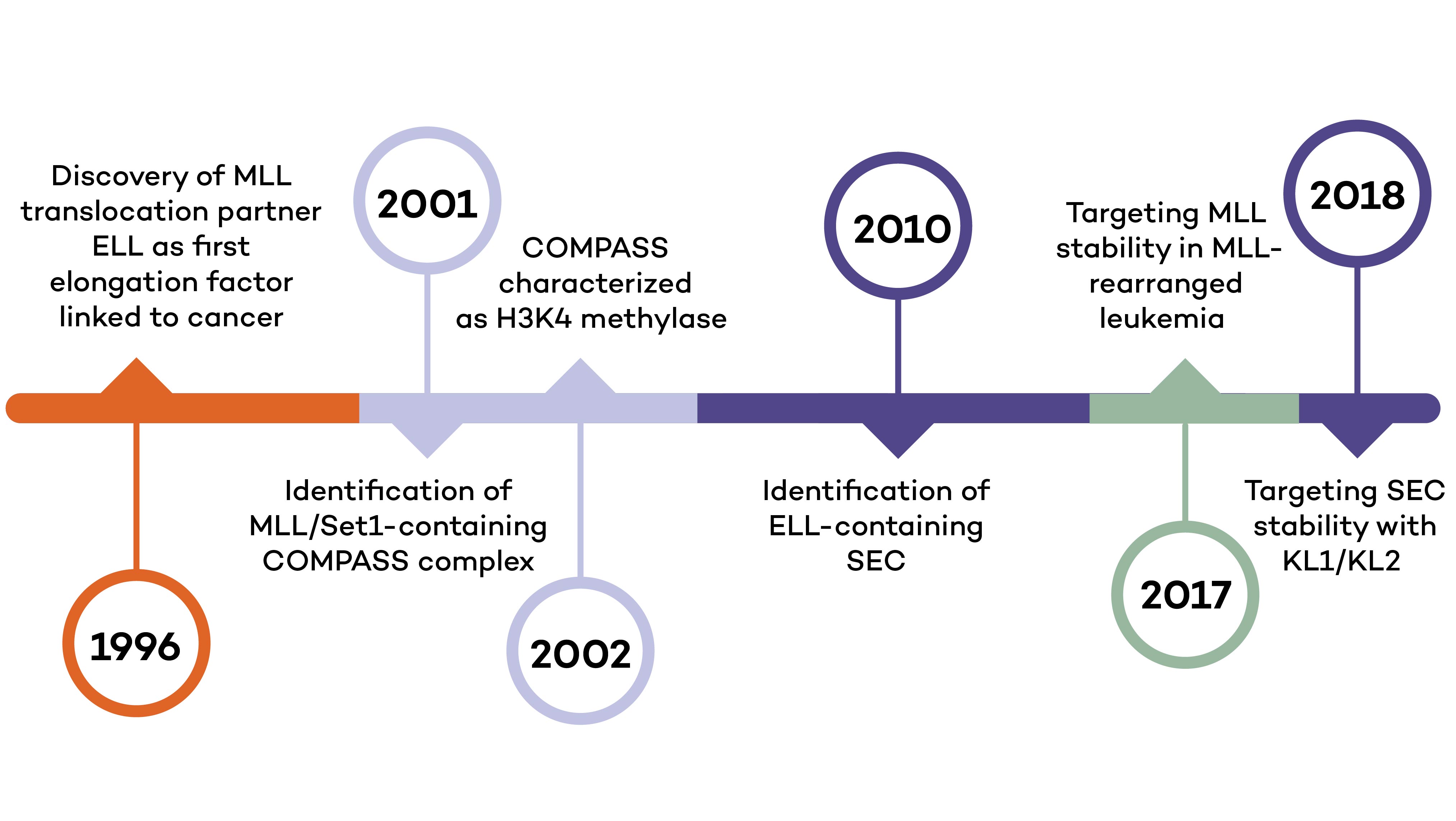
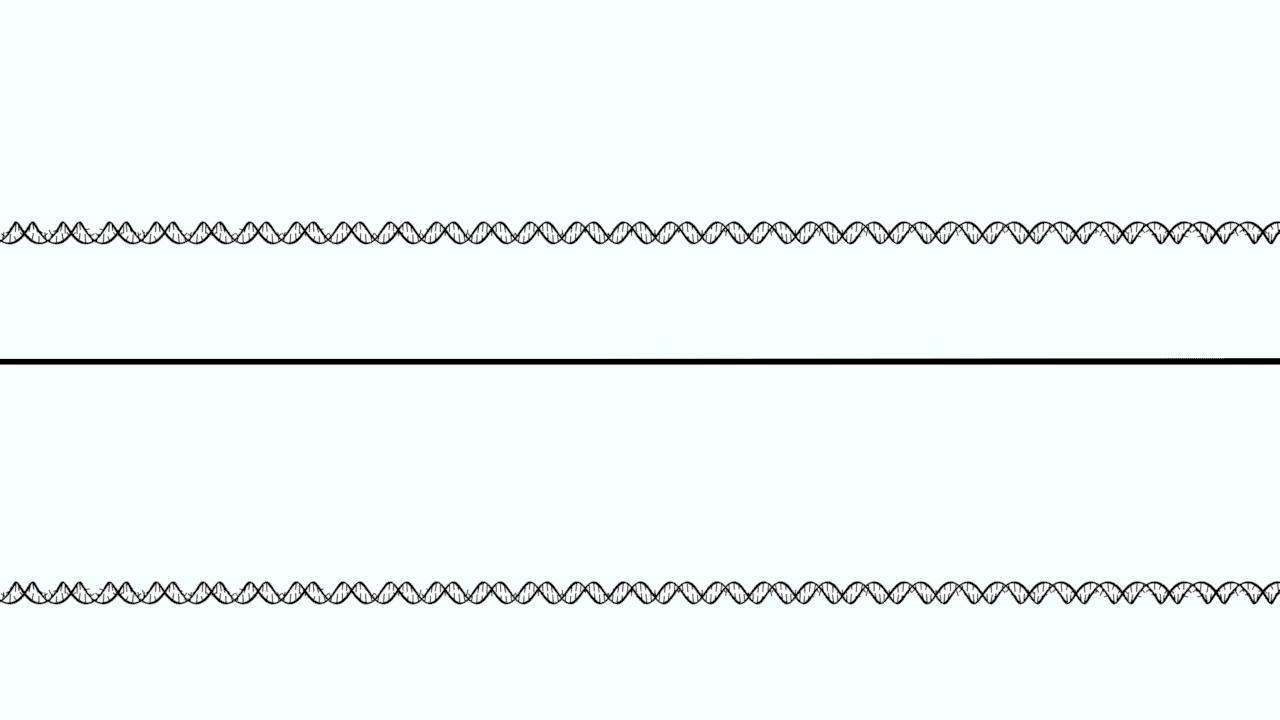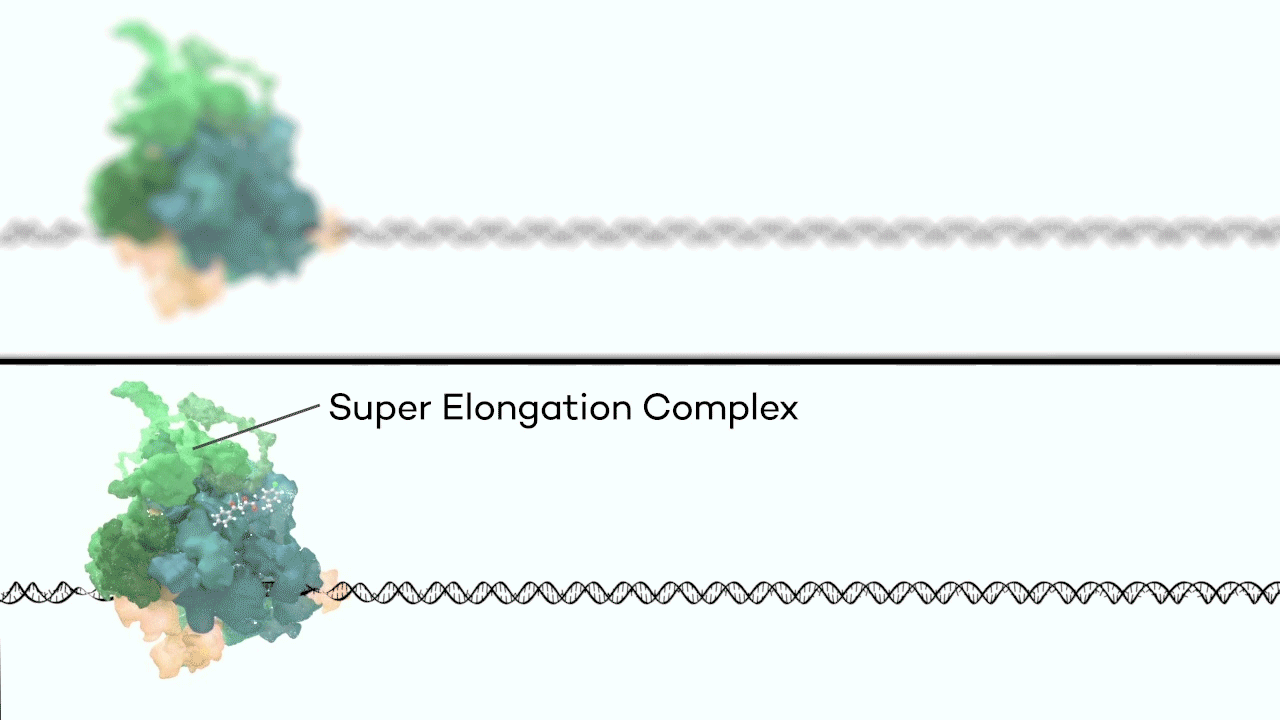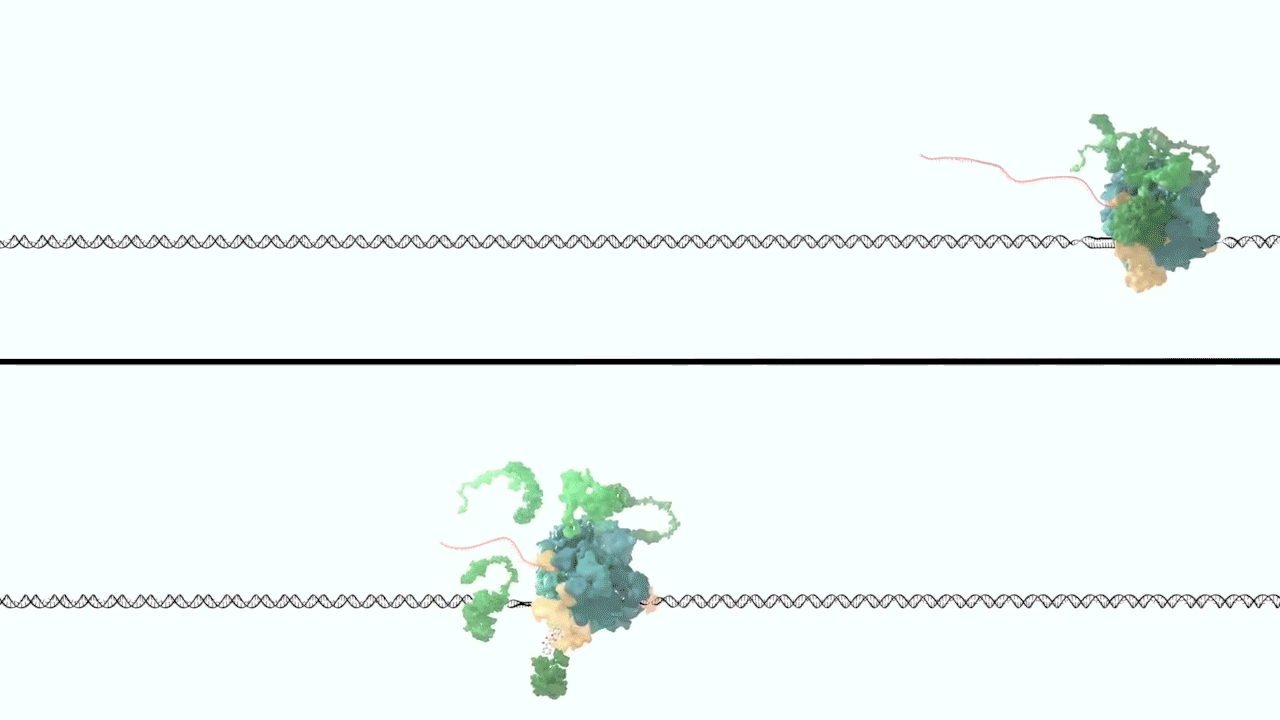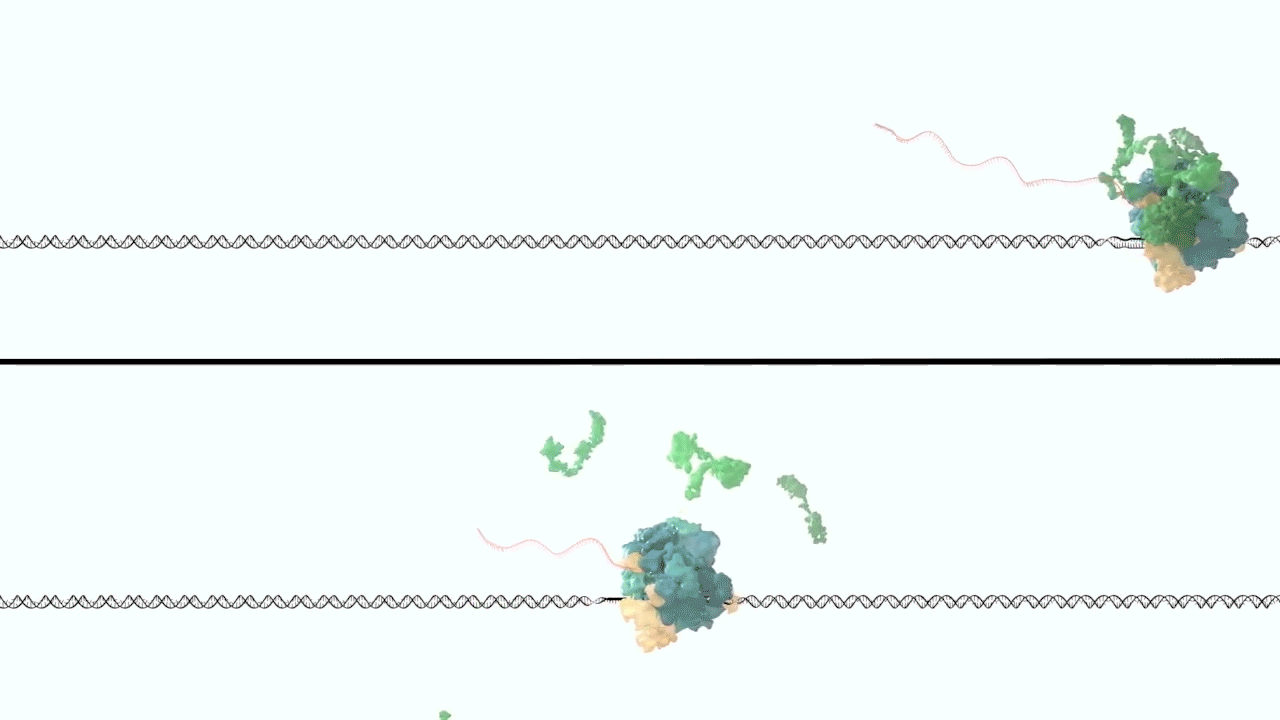
However, it was still unclear to us how the same cancer pathogenesis could result not only from MLL-ELL chimeras, but also MLL translocation-generated fusions with so many other seemingly unrelated factors [Mohan et al., 2010][Shilatifard et al., 2003] Through a decade-long biochemical purification and proteomic analysis of ELL and other common MLL translocation partners we found that, surprisingly, they were all associated with one another in the same complex. We termed this P-TEFb-containing, AFF4-stabilized complex the Super Elongation Complex (SEC). [Chen et al., 2018][Lin et al., 2010]
While these results chart the main course of our past research, their story is only one among the many directions we have explored over the past two decades. However, we find particular interest and satisfaction in discoveries that can continue on into the clinic. After we observed enhanced stability in MLL translocation chimeras relative to their wild type counterparts, we found that ubiquitination activity downstream of IL-1 signaling targets wild type MLL1 for degradation in MLL. In addition, we found that Taspase-1 cleavage unique to wild type MLL1 is facilitated by casein kinase II (CK II). We then demonstrated that either IRAK inhibitors (which block IL-1 signaling) or CKII inhibitors can be used to stabilize wild type MLL so that it can displace dysfunctional MLL chimeras. We also found that MLL cells are sensitive to both IRAK and CKII inhibitors, and that treatment with either inhibitor delays progression and increases survival in a murine MLL-AF9 leukemia model. [Zhao et al., 2019][Liang et al., 2017] IRAK inhibitors are now in clinical trial for AML and MDS, and our results expand the possible clinical indications for this therapeutic direction to include MLL-rearranged leukemias.